Flagellum

The flagellum is a molecular motor (nanomachine) used by microscopic organisms, such as bacteria and protozoans, to propel themselves through an aqueous medium. The bacterial flagellum is a helical filament that rotates like an outboard motor, while the eukaryote flagellum is a whip-like structure that lashes back and forth.
Molecular machines, such as the flagellum serve as powerful examples of the kinds of sophisticated intelligent design that can be found in even the simplest of organisms. ID advocates, such as Michael Behe have helped elucidate the flagellum as irrefutable proof of ID by illustrating the irreducibly complex function of its systems.
Modern physicists look to molecular motors, like the bacterial flagellum, in an attempt to clean methods of designing modern engines in the hope of reaching the functional efficiency evident within the creation.
| “ | According the American Institute of Physics: The structural designs and functional mechanisms to be revealed in the complex machinery of the bacterial flagellum could provide many novel technologies that would become a basis for future nanotechnology, from which we should be able to find many useful applications..[1] | ” |
Bacterial flagellum
The bacterial flagellum is composed of about 25 different proteins, each of them in multiple copies ranging from a few to tens of thousands. It is driven by a rotary motor with a diameter of only 30 to 40 nm. Much like the modern rotary engine, the bacterial flagellum consists of many different parts that carry out different functions, such as a rotary motor, bushing, drive shaft, rotation-switch regulator, universal joint, helical propeller, and rotary promoter for self-assembly.[1] The filament of the bacterial flagellum is a hollow tube composed of protein, and is only 20 nanometers thick. It is helical, and has a sharp bend just outside the outer membrane called the "hook" which allows the helix to point directly away from the cell. A shaft runs between the hook and the basal structure, passing through protein rings in the cell's membranes that act as bearings.[2]
A simplified description of the mechanical features of the flagellum is found in 'Molecular motors of the bacterial flagella', wherein the authors state,
| “ | The bacterial flagellum, which is responsible for motility, is a biological nanomachine consisting of a reversible rotary motor, a universal joint, a helical screw, and a protein export apparatus dedicated for flagellar assembly. The motor is fueled by an inward-directed electrochemical gradient of protons or sodium ions across the cytoplasmic membrane. The motor consists of a rotor, a drive shaft, a bushing, and about a dozen stator units. The flagellar protein export apparatus is located at the cytoplasmic side of the rotor. Interactions between the rotor and the stators and those between soluble and membrane components of the export apparatus are highly dynamic. The structures of flagellar basal body components including those of the export apparatus, being revealed at high resolution by X-ray crystallography and electron cryomicroscopy and cryotomography, are giving insights into their mechanisms.[3] | ” |
The engine can operate at 6,000 to 17,000 rpm, but with a filament attached usually only reaches 200 to 1000 rpm.[Reference needed]. The motor drives the rotation of the flagellum at around 300 Hz, at a power level of 10-16 W with energy conversion efficiency close to 100 %.[1]
Origin
Kenneth Miller, a well-known evolutionary biologist from Brown University, points out that certain subsystems of the bacterial flagellum would still be in working order if other parts were removed. The overall flagellar motility system requires around 50 different types of proteins (and underlying genes to code for them). However, it is quite interesting to note that 10 of these genes and the resulting structure within the flagellar motility system also code for what is known as a type III secretory system (TTSS). The TTSS is used as a toxin injector by some especially nasty bacteria that attack both animals and plants. Therefore, Kenneth Miller argues that it is mistaken to use the flagellar system as an example of a truly irreducibly complex machine since around 40 different parts could be removed from the machine without a complete loss of function. Miller also points out that the majority of the protein parts of the flagellar system have other functions as parts of other systems within bacteria.[4]
What now? Has Miller disproved Behe's notion of irreducible complexity? It sure seems like he has - at first glance anyway. However, what Miller seemingly fails to consider is that the function of flagellar motility is still irreducibly complex regardless of other subsystems functions are or are not maintained with various flagellar system reductions. Without a sizable number of specifically arranged protein parts the function of flagellar motility cannot exist. In fact, all systems of function are irreducibly complex. It doesn't matter if subsystem function is maintained. This is like arguing that the motility function of an automobile is not irreducibly complex because the lights still work even if the engine or tires or drive shaft are removed.
But, what about Miller's notion that the working subsystems can be easily added together to form functions with greater and greater minimum structural threshold requirements? This notion is in fact true up to a point. There are many examples of low-level evolution in action. Miller himself points out a few of these in his book, Finding Darwin's God, to include an experiment by Barry Hall. Hall's experiment is in fact very interesting. What Hall did was delete the genes in a type of bacteria called E. coli that produce a protein enzyme called lactase. Without this lactase enzyme the bacteria could no longer digest the sugar lactose for energy. Hall wondered if these bacteria, if grown in a lactose-rich environment, would evolve the lactase function back again using some other aspect of their gene pool. And, they did! In just a few generations (probably a single generation) the colony of a few billion bacteria evolved a brand new lactase enzyme with just a single point mutation to another gene. This gene product did not have the lactase function before. So, this would seem to be real evolution of a novel function in observable time.
Of course, this is where Miller's descriptions of Hall's experiments end. However, what Hall did next is most interesting. Hall wondered what would happen if he deleted the newly evolved lactase gene. Would the bacteria evolve yet another novel lactase enzyme? Hall carried out this experiment and observed the bacteria for over 40,000 generations - - and they didn't evolve another lactase enzyme despite a very high mutation rate in a large population living in a highly selective environment. Frustrated, Hall referred to this colony as having "limited evolutionary potential".[5]
Now, what is it that limited the evolutionary potential of Hall's bacteria? How is it that such a relatively simply function could not be found by random mutation and function-based selection many times in such a large colony living in such a highly selective environment?
The answer, is found in the expansion of non-beneficial gaps between potentially beneficial genetic sequences as one considers functions with greater and greater minimum structural threshold requirements. As it turns out, evolution proceeds quite easily and very rapidly when it comes to functional systems that only require very few structural threshold requirements or a loss of a pre-existing system of interaction (most forms of antibiotic resistance). Occasionally evolutionary mechanisms produce higher level functions where a few hundred loosely specified amino acid residues are required (lactase, nylonase, etc). However, there are no observable examples of evolution in action produce any novel system of function that requires over 1,000 specifically arranged amino acid residues working together at the same time. There's not one example of evolution beyond this level in all of scientific literature - not one example.[6]
Now, consider that the flagellar motility system requires around 10,000 fairly specifically arranged amino acid residues all working together at the same time. The next closest beneficial "steppingstone" subsystem function is not remotely close at this level of functional complexity. Even if two such subsystems could be put together to make the flagellar system, the odds that only a few residue changes would be required are extremely poor. More likely many dozens of residue changes would be required. The odds of having the needed amino acid residue changes present in one bacteria within a huge population at the same time are extremely remote this side of trillions upon trillions of years of time. Such gaps may not seem like much at first glance, but when considered more closely, the odds of their being crossed in what anyone would consider a reasonable amount of time are pretty much impossible.
In this light, consider that the TTSS system is now thought to have evolved from the fully formed flagellum - not the other way around. In 2000, Nguyen et. al. published a paper in Journal of Molecular Microbiology and Biotechnology titled, Phylogenetic analyses of the constituents of Type III protein secretion systems[7] In this paper the authors argue strongly that the TTSS system evolved from the flagellum. Of course, those like Miller fail to point out such conclusions published in mainstream scientific literature. But such conclusions should have been obvious from the beginning. Which came first? - - a cavefish without eyes or a fish in a pond with eyes? Clearly it is much easier to loose something that was already there than it is to create it in the first place. Remember Humpty Dumpty and all the kings men?
| “ | In The Origin of Species Charles Darwin stated: "If it could be demonstrated that any complex organ existed which could not possibly have been formed by numerous, successive, slight modifications, my theory would absolutely break down." | ” |
Flagellum Assembly
Gallery
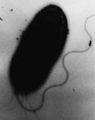
Electron micrograph of H. pylori possessing multiple flagella.
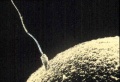
A bacterium with a whip-like flagellum.
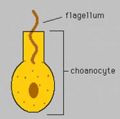
Diagram of choanocyte. The flagella create water currents that bring nutrients into the interior of a sponge.
References
- ↑ 1.0 1.1 1.2 Self-Assembly of Bacterial Flagella by the American Institute of Physics
- ↑ Stoichiometry and turnover in single functioning membrane protein complexes by Mark C. Leake, Jennifer H. Chandler, George H. Wadhams, Fan Bai, Richard M. Berry & Judith P. Armitage, Nature volume 443, pages 355–358, 21 September 2006
- ↑ [1] Molecular motors of the bacterial flagella by Minamino T, Imada K, Namba K. 2008 Dec;18(6):693-701
- ↑ The Evolution of the Flagellum by Dr. Sean D. Pitman May, 2006.
- ↑ Limited Evolutionary Potential Sean D. Pitman M.D. 2006. November 2006.
- ↑ The Steppingstone Problem: And the Limits of Evolutionary Potential by Sean D. Pitman, M.D. March 2004.
- ↑ Phylogenetic analyses of the constituents of Type III protein secretion systems. J Mol Microbiol Biotechnol. 2 (2), 125-144, 2000.
External links
Creationist
- The Bacterial Flagellum by Design and the Universe
- Evolving the Bacterial Flagellum Through Mutation and Cooption By Mike Gene
- The Evolution of the Flagellum: And the Climbing of "Mt. Improbable " by Sean D. Pitman, M.D.
Secular