Hairpin loop
From CreationWiki, the encyclopedia of creation science
Jump to navigationJump to search
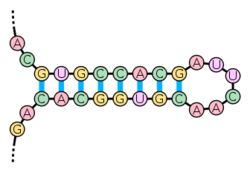
A hairpin loop or stem-loop is a structure that can occur in single-stranded DNA. In this structure, the pairing of intramolecular base pairs occurs.[1] It takes place when two regions of the same molecule, usually forming a palindrome, form pairs to constitute a double helix ending in a loop unpaired. The hairpin loops vary in length from 7 to 20bp.[2] The resulting structure is a building block of many RNA secondary structures. Intrinsic termination is a mechanism in prokaryotes that causes RNA transcription to be stopped. Intrinsic terminators include hairpin loops followed by a run of U residues.[3]
References
- ↑ Ye, Yuzhen; Tang, Haixu (2008). "2-Dynamic Programming Algorithms For Biological Sequence and Structure Comparison". In Mandoiu, Ion I.; Zelikovsky, Alexander. Bioinformatics Algorithms: Techniques and Applications. New Jersey: John Wiley & Sons. p. 19-20.
- ↑ Krebs, Jocelyn E.; Goldstein, Elliot S.; Kilpatrick, Stephen T. (2011). Lewin's Genes X. Boston: Jones and Bartlett Publishers. p. 526. ISBN 978-0-7637-6632-0.
- ↑ Krebs, Jocelyn E.; Goldstein, Elliot S.; Kilpatrick, Stephen T. (2010). Lewin's Essential Genes (2nd ed.). Boston: Jones and Bartlett Publishers. p. 284-285. ISBN 978-0-7637-5915-5.
| |||||||||||||||||