DNA sequencing
DNA sequencing is the process of determining the order of the bases in DNA. Each DNA molecule is made up of four bases: adenine, thymine, cytosine, and guanine. The order of the bases serve as the code to tell cells what to do. There are several ways to sequence DNA. Two popular techniques are the Sanger method and the Maxam-Gilbert method. The Sanger method has set the basis for modern DNA sequencers while the Maxam-Gilbert method has faded away. The two methods have allowed scientists to better understand DNA. Scientists can learn many things by determining the order of DNA. It can lead to advancements in the biomedical field and it could even help prevent genetic diseases. [1]
What is DNA Sequencing?
DNA is made up of four unique bases. The four bases are adenine, thymine, cytosine, and guanine. In a DNA molecule, the order of the bases make up the strands of DNA. The order of the bases serves as a code to tell the cells what to do. DNA sequencing involves determining the order of the four bases in a DNA molecule. [1]
Finding the sequence of DNA can lead to a better understanding in many research projects today. Understanding the DNA sequence in humans can lead to better treatment of genetic diseases. It will also help scientists spot mutations and genetic diseases more accurately and quicker than before. DNA sequencing will lead to many advances in the biomedical industry. Scientists will be able to see what causes certain diseases by looking at the DNA. DNA sequencing might also allow scientists to someday be able to personalize treatments based on the genetic makeup on the individual level. Advances in DNA sequencing have also lead to improvements in the forensic science field. [1]
Sanger Method
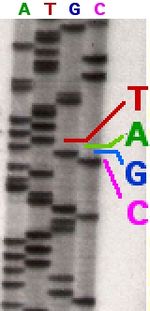
The Sanger method is named after Fred Sanger. Fred Sanger developed the first technique to sequence DNA in the 1970’s. The technique requires the use of four deoxynucleotides: dATP, dGTP, dCTP, and dTTP. The deoxynucleotides cut DNA when they come across a certain base. DATP cuts adenine, dGTP cuts guanine, dCTP cuts cytosine, and dTTP cut thymine. To start the process, primer is added to the DNA to create a DNA template. After the DNA template is created it is put into four separate vessels. After that, one of the deoxynucleotides is added to each of the vessels. The vessels are then heated to help speed up the reaction. The contents are then run on gels through a process called gel electrophoresis. There would be four different lane, one for each of the DNA templates that have deoxynucleotides. Since the deoxynucleotides cut the DNA at certain bases, it would produce fragments that have been cut at each of the possible locations. One can then read the gels from bottom to top to determine the sequence of the DNA molecules. For example, if the band that traveled the farthest was cut by dATP, one can determine that the band was made up of a single adenine molecule. One then looks at the band that traveled the next farthest. Whatever lane the band is in is what base it is. [2]
Maxam-Gilbert Sequencing Method
When the Maxam-Gilbert method for sequencing DNA came out, it was initially the most popular method to sequence DNA. However, as time passed it eventually faded away as the Sanger method grew in popularity. To begin Maxam-Gilbert sequencing, one first needs produce a single-stranded chain of DNA. The strand is then exposed to Phosphorus-32 to mark the DNA with a radioactive tag. Next the strand is exposed to piperidine and other chemicals that cut dimethyl sulfate and hydrazine. Different combinations of the chemicals cut DNA at specific points. One can use these chemicals to cut DNA wherever there is an adenine or wherever there is an adenine or thymine. Other combinations can cut the DNA wherever there is a guanine or wherever there is a guanine or cytosine. The combinations of chemicals are then placed in four test tubes. The first test tube will have the combination of chemicals that cut only adenine. The second test tube will have the combination of chemicals that cut both adenine and thymine. The third test tube will have the combination of chemicals that cut guanine. The fourth test tube will have the combination of chemicals that cut both guanine and cytosine. The fragments produced by the chemicals are then separated out using gel electrophoresis. Like the Sanger method, the gels are then read from bottom to top to determine the sequence of the DNA. Because some of the chemicals can cut both adenine and thymine, one has to look at the bands of the lanes containing the combination of chemicals that can cut both adenine and adenine and thymine. If the band appears at the same place in both the lanes, then the band is adenine. If it only appears in the lane that can cut both adenine and thymine then it is thymine. One has to use this strategy to determine the bands with the guanine and cytosine as well. The Maxam-Gilbert sequencing method lost popularity due to several reasons. The first reason it lost popularity was due to the amount of time it took to sequence a small portion of DNA. Another reason scientists stopped using the Maxam-Gilbert sequencing method was the amount of radioactive material needed to sequence DNA. [3]
DNA Sequencers
Today DNA sequencing is primarily automated through the use of DNA sequencers. DNA sequencers are able to sequence large chains of DNA at a relatively low price. Many DNA sequencers utilize the basic principles of the Sanger method. One example of a DNA sequencer is the technology developed by Pacific Biosciences called Single Molecule Real-Time sequencing. Single Molecule Real-Time sequencing is able to read an average of 15,000 base pairs at a single time at an accuracy of over 99%. This technology allows scientists to sequence much more DNA than ever before at a much higher accuracy. It also makes large sequencing projects a lot cheaper than before.[4]
Human Genome Project
The Human Genome Project was a global research effort to sequence the human genome. The project started in 1990 and ended in 2003. The Human Genome Project sequenced the three billion base pairs to determine the locations of different genes in the genome. The Human Genome Project found that there are around 30,000 genes in the human genome. The completion of the project has been proved to be 99.99% accurate. Determining the sequence of the human genome has helped to improve the biomedical industry. It has helped scientists to spot and cure diseases. The ultimate goal would be for scientists to help treat individuals better based on their genetic makeup. [5]
References
- ↑ 1.0 1.1 1.2 DNA Sequencing What is Biotechnology. Web. Accessed April 29, 2018. Unknown Author. Cite error: Invalid
<ref>tag; name "biotech" defined multiple times with different content Cite error: Invalid<ref>tag; name "biotech" defined multiple times with different content - ↑ Shaffer, Catherine. Sanger Sequencing News Medical Life Sciences. Web. Accessed April 29, 2018.
- ↑ Castelao, Cindy. Maxam-Gilbert Sequencing: What Was It, and Why It Isn’t Anymore Bitsize Bio. Web. Accessed May 6, 2018.
- ↑ Advance Genomics With Long-Read Sequencing, Enabled By Single Molecule, Real-Time (SMRT) Sequencing PacBio. Web. Accessed May 6, 2018. Unknown Author.
- ↑ What is the Human Genome Project? National Human Genome Research Institute. Web. Updated May 4, 2012. Unknown Author.
| ||||||||||||||||||||